
Nuclear factor kB (NF-kB) is a transcriptional regulator that plays a central part in responses to inflammatory signaling not only through Toll-like receptors, but also through TNF receptors and the IL-1 receptor, which we discuss next, as well as in the diverse responses to other signals operating through the TNF receptor superfamily (see section 2–7). It is also essential for responses to signaling through the variable antigen receptors of lymphocytes, which we describe later in the book. We shall see later in this section that although the protein is always called NF-kB, it is a group of related homodimeric and heterodimeric transcription factors that are likely to activate distinct sets of target genes.
Among the molecules induced by NF-kB are cytokines, chemokines, effector molecules of immunity and pro-survival factors (Figure 3-7.1). The pro-survival effects of NF-kB can counter otherwise apoptotic signals coming from cytokine receptors such as TNF receptor I, which we discuss later, and can also protect stressed cells (this can be a factor limiting the effectiveness of cancer chemotherapeutic agents). Mutations that inactivate NF-kB are generally lethal because of the essential role of this protein in cell survival. Partial loss of function causes varying degrees of immunodeficiency: humans with such mutations have variable levels of immunodeficiency and many show poor inflammatory responses and lack some types of antibodies. These symptoms reflect the roles of NF-kB in innate immunity to bacteria (presumably via TLRs), inflammatory gene expression and B cell antigen receptor signaling.
NF-kB is regulated by inhibitory subunits
Figure 3-7.2
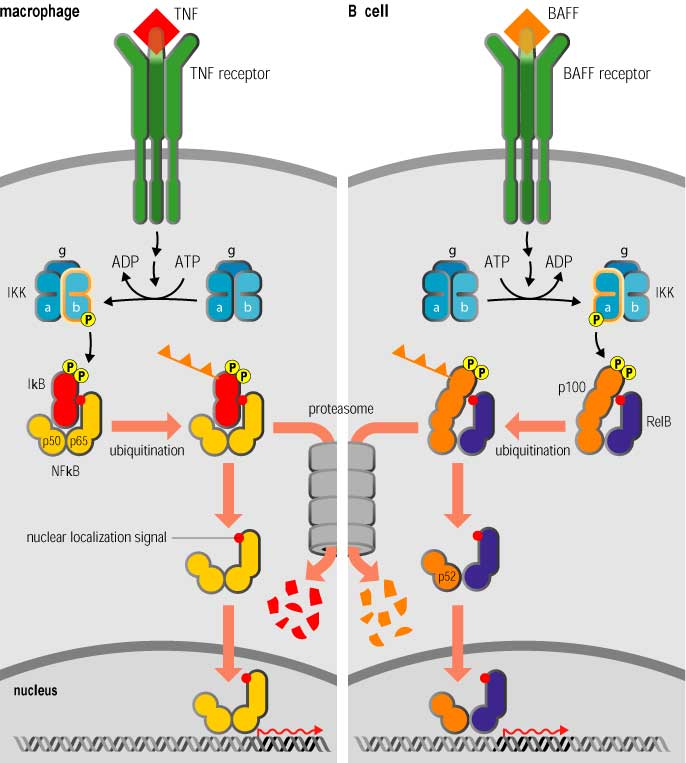
The classical and alternative pathways of NF-kB activation
NF-kB dimers are constructed from five different gene products, all of which have a conserved region called the Rel homology region. The C-terminal domain of this region is responsible for dimerization and I-kB binding, whereas the N-terminal domain is primarily responsible for DNA binding specificity. Three of these subunits, called c-Rel, p65 or RelA, and RelB, also have a transactivation domain; the other two, p50 or NF-kB1 and p52 or NF-kB2, are initially synthesized as much longer precursors containing a regulatory domain (see below). NF-kB family members can form homo- or heterodimers with most other family members, to produce gene regulatory complexes with different properties. For example, the p50–p50 dimer is a transcriptional repressor rather than activator.
The classical pathway of NF-kB activation is illustrated in the left panel of Figure 3-7.2, which shows an inflammatory response operating through a heterodimer of p50 and p65. NF-kB dimers are held in the inactive state by a family of inhibitors called I-kB. Receptor signaling leads to activation of a multisubunit I-Bk kinase (IKK) complex which phosphorylates I-kB on two key serines. Phosphorylation of I-kB marks it for degradation by the ubiquitin pathway (Figure 3-7.2), so that the NF-kB dimer is liberated to translocate to the nucleus, bind DNA and activate transcription.
It is essential that the inflammatory actions of NF-kB are switched off once the inflammatory signal ceases, and because the inhibitor I-kB is degraded on NF-kB activation, this means new I-kB must be synthesized. There are three main members of the I-kB family, two of which, IkBß and I-kBe, are synthesized constitutively and reestablish NF-kB inhibition on cessation of signaling with a relatively slow time course. Synthesis of the third, I-kBa, is under the control of NF-kB itself, and it is therefore produced in response to signaling: it enters the nucleus on synthesis, binds to NF-kB and shuttles it back to the cytoplasm via a nuclear export signal, switching off NF-kB action with a very short delay, thus making NF-kB activity self-limiting.
Some NF-?B heterodimers are self-inactivated
Although the mechanism illustrated on the left in Figure 3-7.2 is the usual pathway of NF-kB activation, there is an alternative pathway in which the NF-kB dimer is held in the inactive state by the extended C-terminal domain of the precursor form of one of the subunits. Both p50 and p52, as we have mentioned, are synthesized as longer precursors (p105 and p100 respectively). The extended C-terminal domains of these precursors are structurally homologous to I-kB and have the same function. While the p105 precursor is believed to be constitutively processed to give p50, p100 is not, and serves as a regulatory partner in the NF-kB heterodimer. This is seen in the survival pathway of B lymphocytes, which contain p100–RelB heterodimers that are held in an inactive state by the C-terminal domain of p100 until they are activated by the TNF-family molecule BAFF (B-cell activating factor). As shown in the right-hand panel of Figure 3-7.2, receptor activation leads to proteolytic processing and degradation of the inhibitory C-terminus of p100 and liberation of the remainder as the transcriptionally active p52–RelB complex.
Definitions
I-kB: inhibitors of NF-kB, existing in multiple isoforms, all of which contain ankyrin repeat structures that mediate the interaction with NF-kB.
I-kB kinase (IKK): a complex consisting of two related kinase subunits, called IKKa and IKKß, and a scaffolding subunit, IKKa or NEMO. IKKß and IKKa are necessary for responses via the classical pathway, but not the p100–RelB pathway, which only requires IKKa. Conversely, IKKa is dispensible for the classical pathway.
nuclear factor kB (NF-kB): dimeric DNA binding proteins, originally described for their binding to the B site in the Ig a intronic enhancer.
proteasome: a large multisubunit complex of proteases in the cytoplasm, which primarily degrades poly-ubiquitinated proteins.
Rel homology region: a 300 amino acid long homology region in all five NF-kB types. This homology forms two immunoglobulin-like ß-sheet sandwich structures, the N-terminal of which is responsible for most of the specificity of DNA binding and the C-terminal of which is responsible for dimerization and binding to I-kB.
References
Claudio E, et al.: BAFF-induced NEMO-independent processing of NF-kappa B2
in maturing B cells.
Nat Immunol 2002, 3:958-965. [PubMed Abstract][Publisher Full Text]
Ghosh S, Karin M: Missing pieces in the NF-kB puzzle.
Cell 2002, 109:S81-S96. [PubMed Abstract][Publisher Full Text]
Hoffmann A, et al.: Genetic analysis of NF-kB/Rel transcription factors defines
functional specificities.
EMBO J 2003, 22:5530-5539. [PubMed Abstract][Publisher Full Text]
Hoffmann A, et al.: The IkB-NF-kB signaling module: temporal control and selective
gene activation.
Science 2002, 298:1241-1245. [PubMed Abstract][Publisher Full Text]
Jacobs MD, Harrison SD: Structure of an I-kBa/NF-kB complex.
Cell 1998, 95:749-758. [PubMed Abstract][Publisher Full Text]
Puel A, et al.: Inherited disorders of NF-kB-mediated immunity in man.
Curr Opin Immunol 2004, 16:34-41. [PubMed Abstract][Publisher Full Text]