|
|

|

|
|

|
From August 2009 to July 2010 I am a National Research Council postdoc at the US Environmental Protection Agency. I have been working on various methods for spatiotemporal data, including predictions related to the primary and secondary ozone standards. To this end, I started an R package, spt, which is a set of S4 classes for spatiotemporal data. It's still in alpha. It's hosted at R-forge.
I was in Costa Rica from July 2008 to July 2009 to support my spouse's Ph.D. field research. While there I defended my thesis, did some contract work and some volunteering. Before the economy tanked, I was a software engineer at Zilliant from June 2007 to October 2008, with limited research roles in the pricing industry. From what I understand the name Zilliant came from a party where the founder (whose last name allegedly started with a "Z") spoke with an inebriated partygoer who said, Z.., you're brilliant, you're Zilliant... Don't know how true that is...
From May 2006 to June 2007 I was a contract statistician (official title was "computational statistical geneticist") at GlaxoSmithKline in Research Triangle Park, NC.
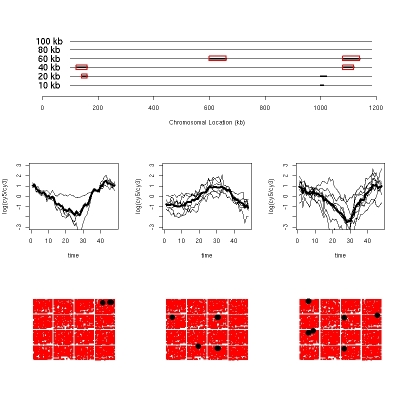
Before December 2005, I worked for Rudy Guerra dealing with high throughput biological data. Our collaborators included biologists and physicians at MD Anderson Cancer Center, Baylor College of Medicine and the University of Texas Medical Branch. We are also participating in the Critical Assessment of Micrarray Data Analysis (CAMDA) 2004 competition working with malaria (Plasmodium falciparum) micrarray and sequence data. Our group (Myself, Chad Shaw, Rudy Guerra, Josue Noyola-Martinez, Garrett Fox, Marie Stevens, Mike Gustin, Dennis Cox and David Scott) had both of our extended abstracts selected to give presentations, so we were in North Carolina November 10-13, 2004. We split a first prize for best presentation, and I have included one of the plots from that paper above, showing (top) regions of the chromosome with significant regions of correlated RNA transcripts (middle) the correlated transcripts from those regions and (bottom) the locations of the probes on the chip (as a check to make sure that there was no confounding due to printing location).
In spring 2004 I was a VIGRE graduate student working for Rudy Guerra, with a loose focus on modeling biological networks. In fall 2003, I was a resource for the VIGRE junior learning seminar, and taught a section of CAAM 210 in spring 2004.
Formerly, I was a research assistant for Dr. David W. Scott from
August 2001 to May 2003, working on his Digital
Government Quality Graphics grant. Please visit our research
site for more information.
I briefly worked as a research assistant for the mouse atlas project. One of the goals of this project is to efficiently store and search in-situ gene expression from the mouse brain. The mouse brain is sliced into pieces a couple of cells thick, and stained so that cells expressing a certain gene are colored according to the amount of gene expression. There's a little more to the story, but this is close enough. My job was to help design the search algorithm (ie find me another gene with the same spatial expression as this one) and provide p-values for the quality of gene expression matches.