|
|
Marina Vannucci
Noah Harding Professor and Chair of Statistics, Rice University
This website contains links to codes developed by Dr. Vannucci and her students and collaborators, together with some of the datasets used in their publications.
GitHub Repositories
SSBVS - Stochastic Search Bayesian Variable Selection
- Matlab code for Multivariate Linear Regression Models.
Reference paper: Brown, P.J. et al. (2002). Bayes
model averaging with selection of regressors. JRSS, Series B, 64(3), 519-536.
- Matlab code for Multinomial Probit Regression Models.
Reference paper: Sha, N. et al. (2004).
Bayesian variable selection in multinomial probit models to identify molecular signatures of disease stage.
Biometrics, 60(3), 812-819.
- Matlab code for Accelerated Failure Time Models.
Reference paper: Sha, N. et al. (2006).
Bayesian variable selection for the analysis of microarray data
with censored outcome. Bioinformatics, 22(18),
2262-2268.
- Datasets:
R Packages in CRAN
Other Code
- Matlab code
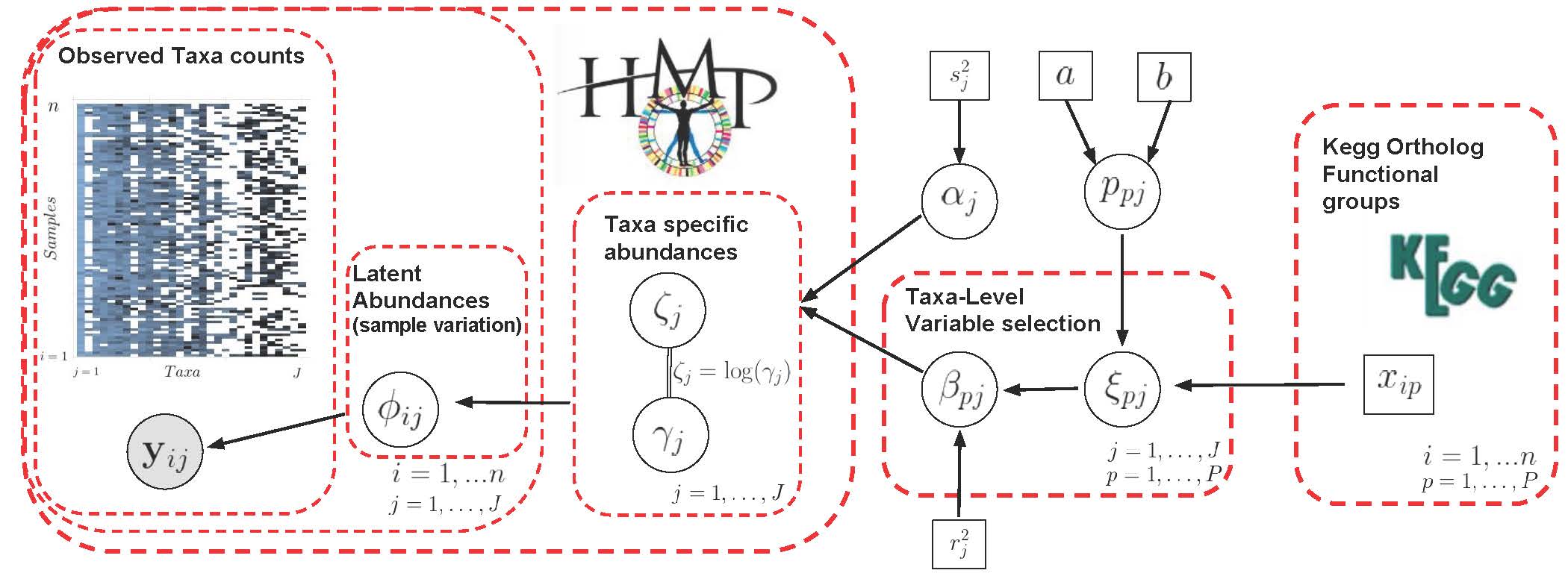
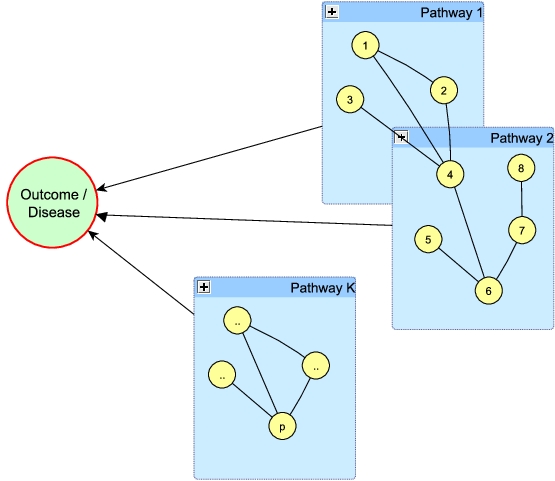
Reference paper: Stingo et al. (2011).
Incorporating Biological Information into Linear Models: A Bayesian
Approach to the Selection of Pathways and Genes. Annals of Applied
Stats, 5(3), 1978-2002.
- Matlab code
Reference paper: Stingo et al.(2013).
An Integrative Bayesian Modeling Approach to Imaging Genetics.
Journal of the American Statistical Association, 108, 876-891.
Dataset: Processed fMRI and SNP data
on 210 subjects from a study on schizophrenia
provided by the MCIC consortium (Gollub et al., Neuroinformatics,
11(3), 367-388, 2013).
- Matlab code
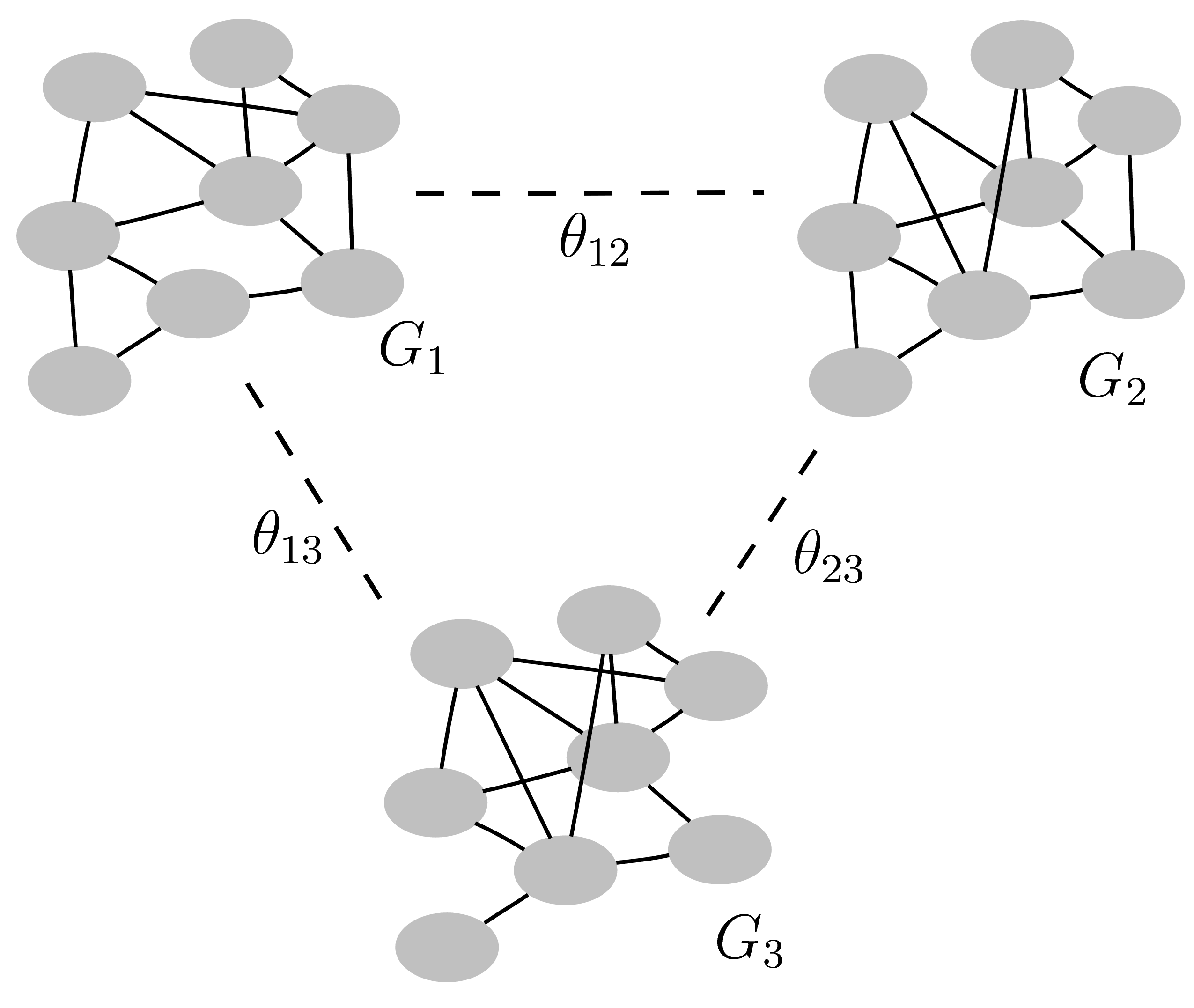
Reference paper: Peterson et al.(2016).
Bayesian Inference of Multiple Gaussian Graphical Models.
Journal of the American Statistical Association, 110, 159-174.
Protein Structure Prediction
-
Web app for Bayesian Modeling of Protein Primary Sequence for
Secondary Structure Prediction (paper).
- Software for
A Dirichlet process mixture of hidden Markov models for protein structure prediction
(paper).
|
|

